


The epoxidation of trifluoromethylpropenol.
Molden can animate reaction paths from Mopac/Ampac
.arc files, Mopac IRC output files (provided
the mopac keywords; xprio and large were used to construct it),
XMOL
cartesian format or Gamess/Gaussian optimise runs. (See Input Formats)
In both cases the energy value per point is extracted as the first
real number from the title line.
If you have properly set up Molden as as a
Chemical MIME type viewer, then click
Here to view the racemization of hexahelicene.
Now first select Solid and Shade from the
Molden Control Window.
and rotate the molecule to get the following view:
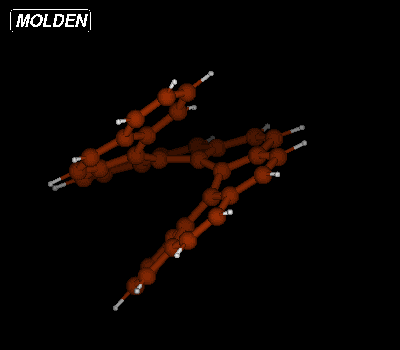
Now select Geom. conv. from the Molden Control Window. The Geometry Convergence window will now pop up:
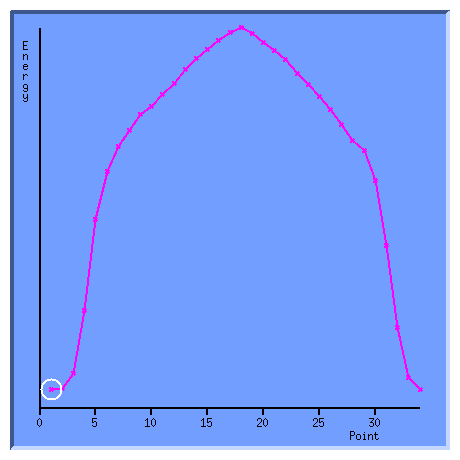
This window represents a plot of the energy against the geometry points. The currently displayed geometry is denoted by a white circle. Putting the cursor over the a particular point in the curve will temporarily display it's energy. Clicking on that point in the curve will select the associated geometry for display in the main window. You can get rid of this window by clicking in it with the third mouse button or selecting Geom. conv. from the Molden Control Window again. But for now leave it on the display. We can now animate a walk over the reaction path by selecting Movie from the Select Point: section of the Molden Control Window:
You can also do this stepwise by using the First and Next button of the Select Point: section of the Molden Control Window.
Finally, if you have properly set up Molden as as a Chemical MIME type viewer, then click Here for a second example.