

| 
|
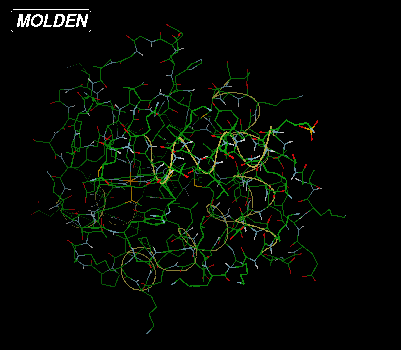
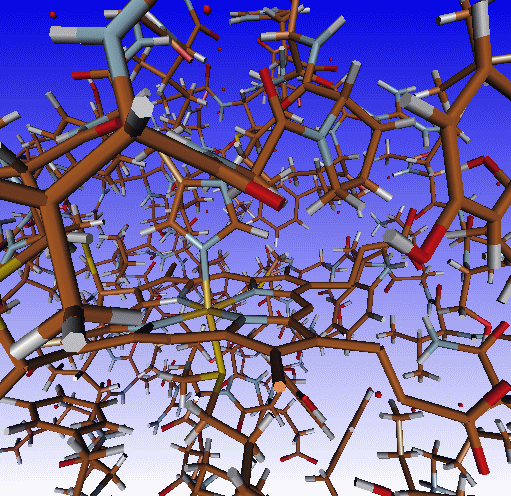
Above are depicted on the left the standard X-windows display (molden
executable, Draw mode Solid off) and on the right the
OpenGL display (gmolden executable, Draw Mode Solid on -->
Ball & Stick) 
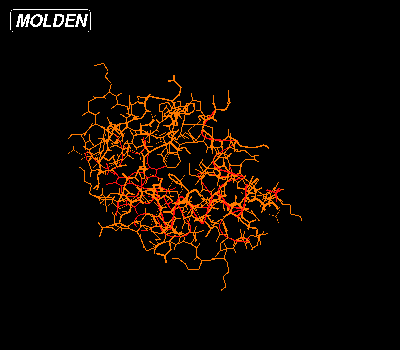
If you have properly set up Molden as as a Chemical MIME type viewer, then click Here to view the Cytochrome C from Baker's yeast entry (1YEB) from the Protein Data Bank (PDB). The default view for PDB files is the following:
| 
|
In this view all amino acids are colored orange, the substrate is colored red and the helices are also colored red. To get to the first view select StickColor and Shade from the Molden Control Window:.
In the StickColor Draw Mode all atoms are colored by their specific atom color. Now lets return to the default Draw Mode, by selecting StickColor again. By clicking BackBone only the backbone atoms are displayed. And now the Add to BackBone: section becomes active.
Now select Hetatm from the Add to BackBone: section, select "HEM", select a color and the heme will also be displayed in the chosen color.
When moving your mouse over the backbone, a label will appear for the closest amino acid. When the label is visible, click with the first mouse button on the label and a popup menu will appear. Select Switch On from this menu and the amino acid will be displayed:
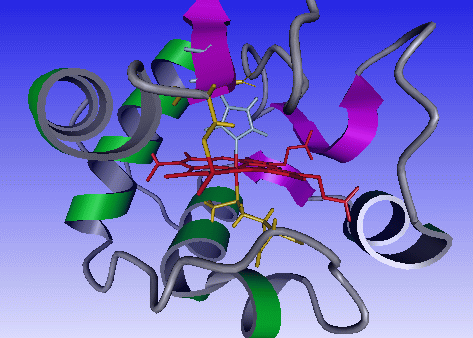
When moving your mouse over the heme, the heme label will appear, click with the first mouse button and select Center. The center of rotation will now be
the heme atom you clicked on.
You could also have chosen Add to surface and the follwing window would appear:
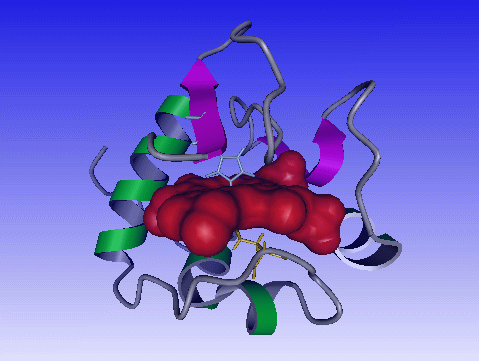
Click color, select a color and click Create Surface. Now the heme will depicted as surface.
You can also create a solvent accessible surface for the whole protein (well amino acids). In the molden control window click the surface icon and select Solv. Acc. Surf. --> Elec. Pot. Mapped:
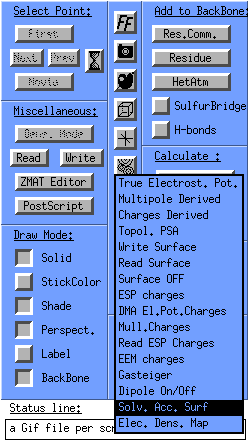
And you will get the following display:

You can manipulate the surfaces by clicking on the color pallete icon and selecting surfaces:
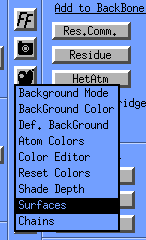
The Surfaces window will appear:
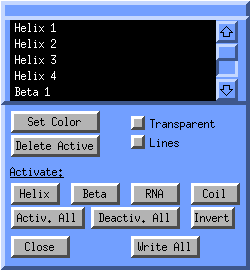
In the listbox all surfaces are listed, such as the helices, strands, coils,
the heme surface and the solvent accessible surface. All current surfaces
are active, (white text on black background). If you want to change the color
of one particular helix, click on its corresponing entry in the listbox.
It will now becoming deselected/inactive and is nolonger displayed.
Click on invert selection, click on Set Color,
select a color and finally click on Activ. All.
Now the particular helix has your own customized color.
You can save complicated scenes by clicking Write All. A molden.ogl will be written. You can load such a scene back into molden by clicking on
its corresponding entry in the Molden File Select window (Read button in the Molden Control Window).
Now if we click on the Res.Comm. (Residue Command) button in the Add to the Backbone section of the Molden Control Window, a query box will pop up.
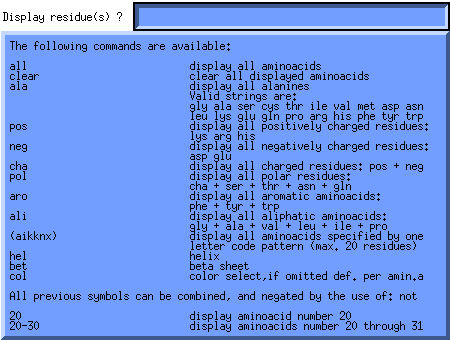
This query box enables the user to switch on types, ranges and patterns of amino acids, helices and beta strands and optionally choose their color. Now lets switch on the alpha helices and color them green, by first putting the cursor in the Residue Command Query Box then typing "hel col" and hitting return. A Color Selection Box will now pop up. Select a the light green color by clicking on it. The molecular display will now look like this:
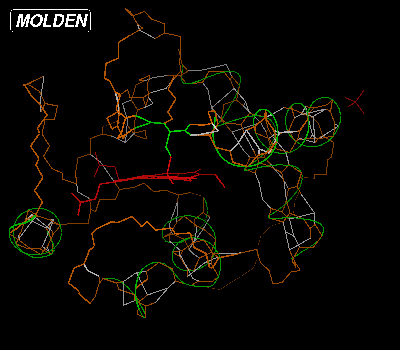
If we would want to switch on all aromatic amino acids, we would type "aro" in the Residue Command Query Box:
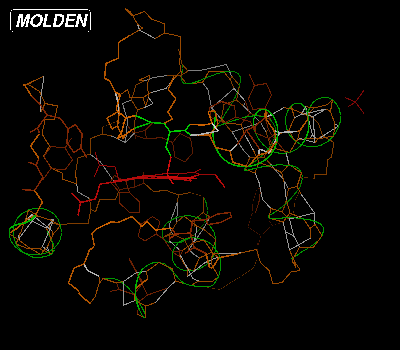
You clear the backbone from amino acids by typing "clear" in the Residue Command Query Box. Then if you type "22-23" and hit Return or Enter, type "9-20 col" and hit Return or Enter, the Color Selection Box will pop up again. Select the blue color:
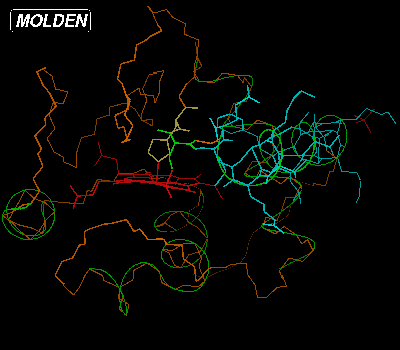
We have now colored the amino acids making up one of the helices blue and lighted up two amino acids connected to the heame, using their default color. If we want to see the active amino acids labelled by type and number, we should click on the Label button from the Molden Control Window. We can have a ball and stick view of the molecule at any time by pressing Solid from the Molden Control Window, mind you the Solid + Shade view will be very slow for proteins in the molden executable, but not for the OpenGL gmolden executable. The display would then look like this:
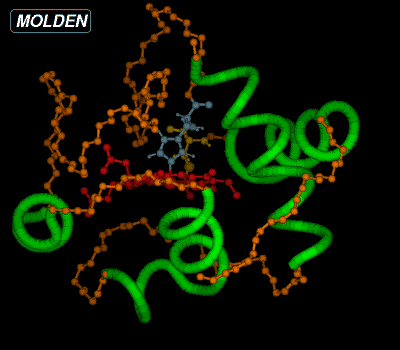
| 
|
We can make a postscript picture of this view by pressing PostScript from the Molden Control Window. A popup box will appear. Put your cursor in this window, type the filename and hit Return or Enter. The resulting postscript file can be viewed here and the color postscript (See Molden command line flags) version here. The lastest version of molden has an improved Color Postscript rendering. (See also as GIF Image).
Finally the perspective Draw Mode can be used to step through large proteins.
Commandline options pertaining to PDB visualisation:
-f PDB: build connectivity from cartesian coordinates
VERY SLOW
-g PDB: always calculate Helix/Sheet information
Default: Take the information from the PDB file.
-P PDB: treat all input files as PDB files
Default: PDB files are recognized when the first line
is: HEADER .....
For use with DNA/RNA:
-hoff switch of hydrogen bonds
-hdmin x mininum hydrogen bond distance (Ang)
-hdmax x maximum hydrogen bond distance (Ang)
-hamin x mininum hydrogen bond angle (Degrees)
-hamax x maximum hydrogen bond angle (Degrees)