

One of the new Molden features is its ability to edit protein structure and to optimise protein models with the help of the Tinker package.
Molden allows you to create proteins from scratch and the ability to modify existing proteins. Internally both creation and editing of proteins is accomplished via the the Z-Matrix Editor. However for the use with proteins the standard Z-Matrix Editor shows to much information. That is why Molden allows you to switch between a protein oriented representation of the Z-matrix and the full Z-matrix by means of a toggle button.

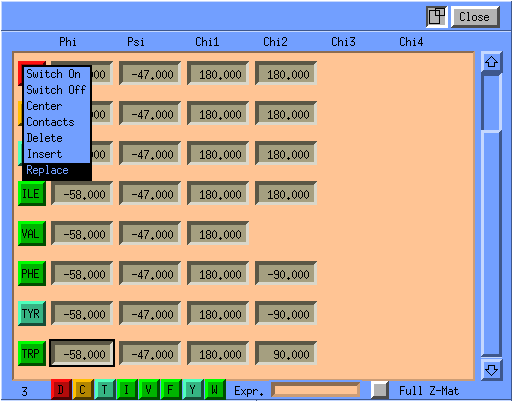
The protein oriented representation of the Z-matrix is composed of a list of the amino acids making up the protein, with each amino acid there is a button, which when pressed will let you manipulate this amino acid. This can range from just switching it on for display to replacing it by an other amino acid. Other editing functions include delete and insert. In addition there are for each amino acid editible fields for the Phi,Psi and Chi angles associated with them. When you click on an amino acid in the structure its line will become the first line in the Z-matrix window. On lower part of the window there is the one letter code equivalent of the current window of eight amino acids. It also has a editable field for specifying a small stretch of one letter code which when found will position the amino acid window to start at the first of these residues.
The Z-matrix editor has a button called Substitute atom by Fragment, one of its options is Sequence, when selecting this the Build Sequence Window will pop-up:
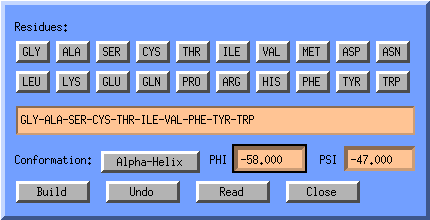
This window allows you to specify parts of a sequence, and lets you select
the Phi and Psi angles associated with it. You can used
predefined values for Alpha-Helix and Beta Strand. This
window builds with respect to previous builds, so you can sequentially add
parts of the sequence.
When you read in a PDB file, a Mol2 file, or a Tinker XYZ
file with either the amber or charmm forcefield applied, molden will ask you
wether you want to create a Z-matrix for this protein when you click the
ZMAT Editor button.
Via the interface to the Tinker package molden allows you to optimise
your protein/polypeptide. First you get rid of the Z-matrix editor window,
by clicking the ZMAT Editor button. Then you popup the force field
window by clicking the  icon in the Molden Control
Window.
icon in the Molden Control
Window.
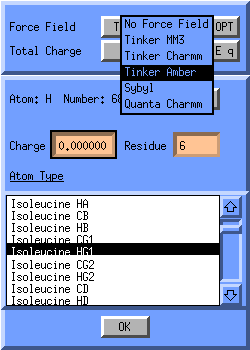
Now you have apply force field typing. Click on the button next to the text Force Field and select either Tinker Charmm or Tinker Amber. Inspect the force field types per atom by switching on the labeling, click the Label button in the Molden Control Window and select ForceF.Type. To start Tinker click on the OPT button. Now the Tinker Preferences window will pop up.
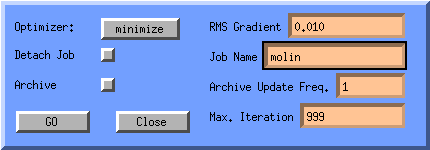
This window will let you choose the Optimizer used by tinker, Newton
is the best but is only feasible for small peptides (<15 res.), minimize
and optimize are suitable for proteins, the dynamic option
does work currently. The Xtinker option is for small crystal
minimisations, and requires a special adaptation of tinker called Xtinker.
The RMS Gradient specifies the root mean square gradient at which
the optimisation is complete.
You can opt for real time on screen optimisation or optimisation in the
background. You can specify that you want an archive file (Jobname.arc),
which molden can read back in and animate via the Movie button.
The Job Name itself can be supplied via an editable text field.
Click GO to start Tinker. Now molden will setup the files needed
to run Tinker, after doing so it will prompt you to go on, this enables
you to customize the files jobname.run and jobname.key.
After a successful run the following files will be created:
jobname.xyz The structure in tinker xyz format
jobname.run The csh script that runs tinker
jobname.log The log file, look for tinker error messages here
jobname.key This file specifies which tinker parameter file is being used
jobname.arc The archive file containing the intermediate and final
structures