

Pharmer is an open source program developed by David Koes from the Camacho Lab in the Department of Computational and System Biology at the University of Pittsburgh. Pharmer powers ZINCPharmer, an online pharmacophore search engine for a multi-conformer library of the ZINC database.
A pharmacophore is a spatial arrangement of functional groups or atoms, together with their respective distances (and tolerances) to each other, that a small molecule must satisfy in order to be active against a receptor protein.

When the pharmer executable is found (default /usr/local/bin) the option
Pharmacophore Editor will become available from the "Surface" icon
![]() (7th from the top of the central icon column).
(7th from the top of the central icon column).
This will run pharmer to create the annotation of your molecule. Pharmer will create a list of features for you to choose from that you think is essential for interaction with the target protein. Below you will find an image of the annotation of the raloxifen molecule.
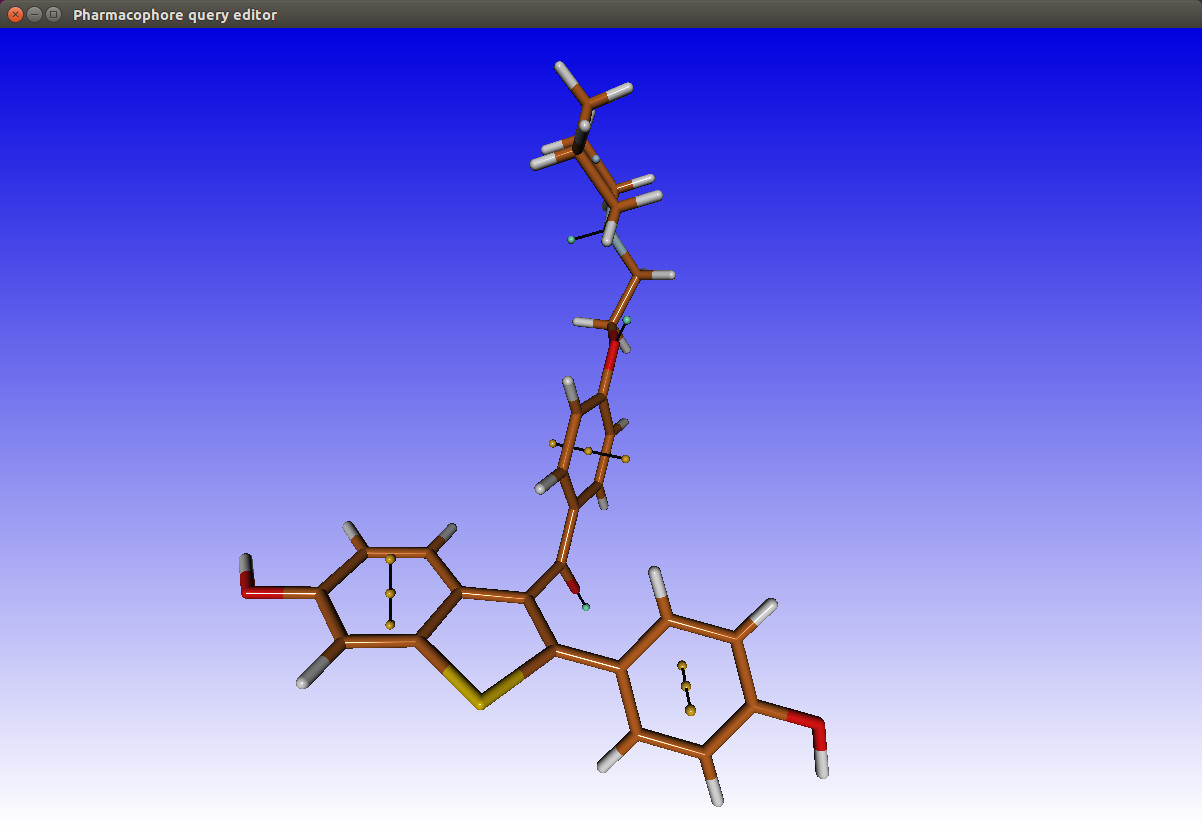
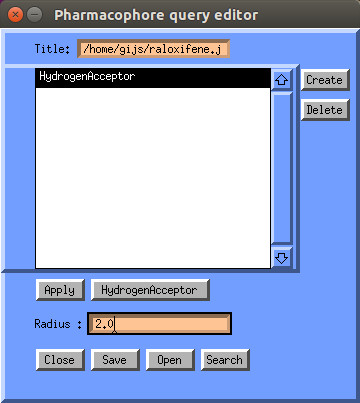
From the pharmacophore Editor window you will be able to add features by
clicking the "Create" button, after you have first clicked the feature in
molecule display.

A selected feature stores the associated coordinates, you can then select other features available in the annotation with the same coordinates. (sometimes multiple features are available with the same coordinates eg: aromatic and hydrophobic). You can also adjust the radius, which defines a tolerance sphere around the feature.
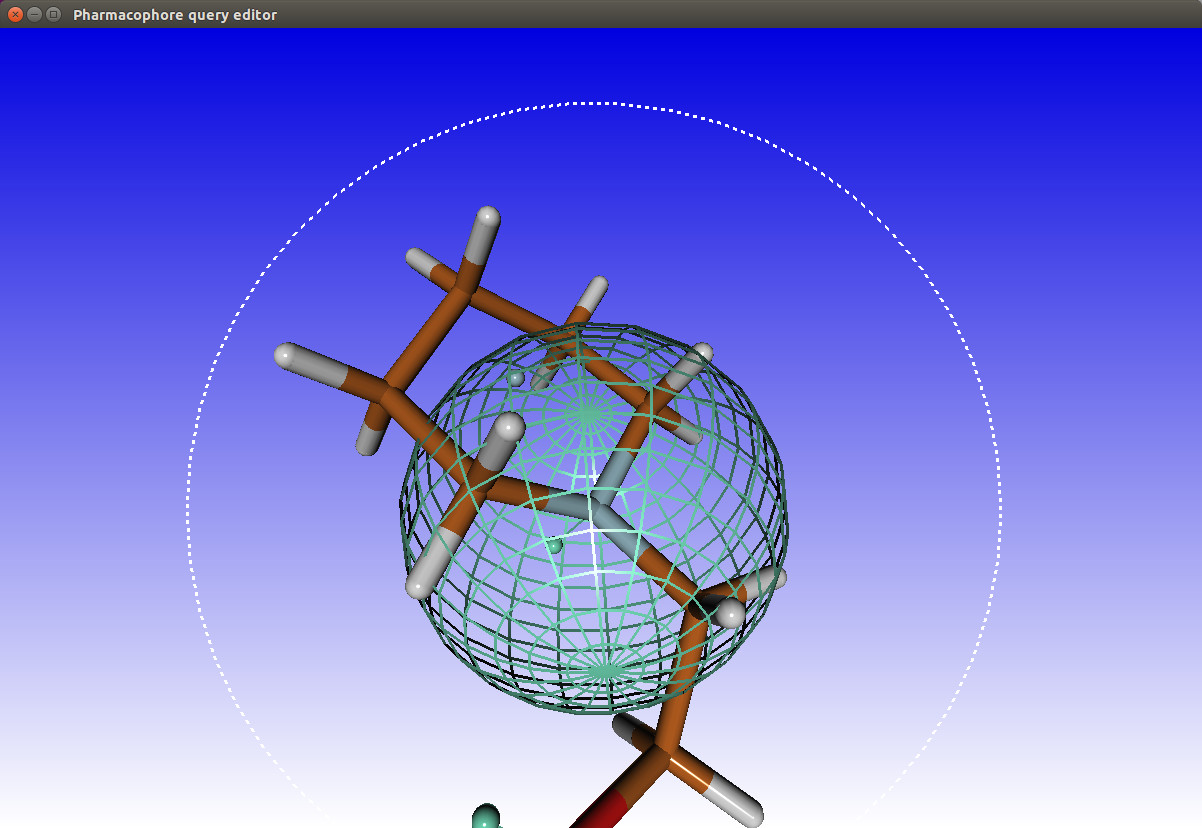
When you are done with selecting features you can save the selection by clicking the "Save" button. You can also restore previous selections by clicking the "Open" button. Selections are stored in the json format.


You are now ready to search a database with this query.
In Molden
database files are presumed to come in the form of a .sdf file
or .mol2 file. Pharmer expects the database in a .sdf file
format.
When clicking "Search", the Pharmacophore Search Database window
will open.

This allows you to select a .sdf database file (Open button). You can then create a Pharmer Database (index) by clicking "Create DB Index". This will create a directory with the same directory name as the original filename of the .sdf file, but with the .db extension.
Finally, clicking the "Search" button in Pharmacophore Search Database window will search the Pharmer database
with the stored .json query and return the results in the .sdf file specified
in the "Results:" query box (default: phrm.sdf).
You can inspect the results by reading the results file into molden.
The molden5.9 version comes with a full distribution for windows, Linux (64 bits ubuntu) and MacOSX (64 bits). The full distribution contains all the files necessary to run the gmolden executable, as well as the openbabel, pharmer and open3dqsar executables, with required auxilliary files.