

Open3DQSAR
is open-source package written by Paolo Tosco and Thomas Balle from
the Department of Drug Science and Technology, University of Turin and
Faculty of Pharmacy, University of Sydney respectively.
Open3DQSAR uses molecular interaction fields (MIF) to calculate descriptors
on each point on a 3D grid surrounding a set of pre-aligned molecules, supplied
in the form of an .sdf file. The alignment should be of the bioactive
conformation of the molecules, which creates a sort of catch 22 situation,
since Qualitive Structure Activity Relationships
(QSAR) is a ligand-based approach which is typically used when there is
no protein 3D structure information available.
The other prerequisite of this method is a measured activity for each compound
in the set of aligned structures, supplied as a text file containing the
activity of each compound, each compound on a separate line.
Each of these descriptors can contribute to the activity (increase activity)
or negatively impact the activity (decreased activity).
For each grid point we get two descriptors. One for each component of the
molecular field: van der Waals and Electrostatic fields.
For the van der Waals field each descriptor is colored green if it
contributes to the actvity and yellow if it negatively impacts the activity.
For the electrostatic field each descriptor is colored red if it
contributes to the actvity and light blue if it negatively impacts the activity.
We can now create surfaces that go through those points on the 3D grid, that
contribute positively or negatively to the activity by a certain measure.
For each field (the van der Waals or electrostatic field) we use a different
color if it contributes positively or negatively to the activity.
Green and yellow for the van der Waals field.
Red and Blue for the electrostatic field.
You will find the new option Open3DQSAR under the
![]() "Surfaces icon"
(the 7th icon from above middle column).
"Surfaces icon"
(the 7th icon from above middle column).
When clicked, this will pop up the QSAR interface window.
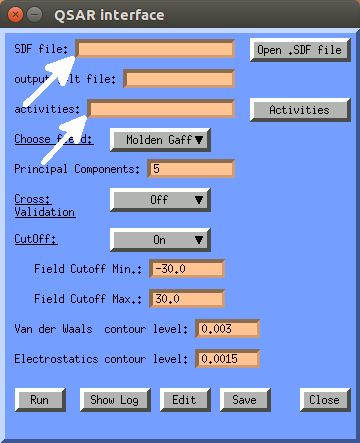
Here the minimum inputs are:
and

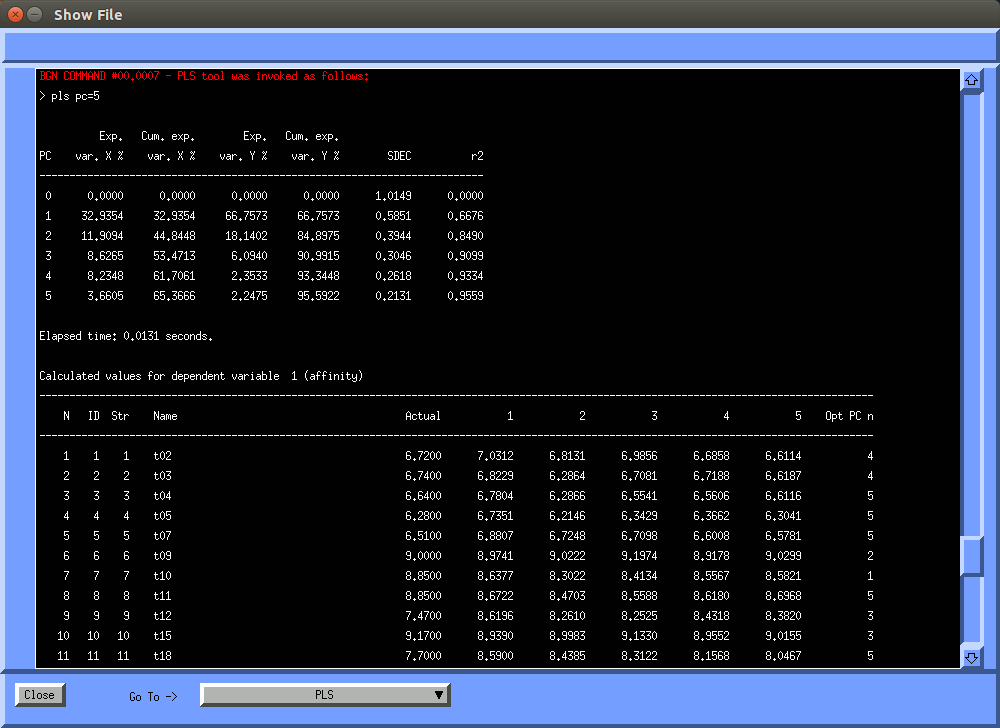
The "Edit" button pops up the "open3dqsar edit commands" window, that shows the list of commands being sent to open3dqsar. When you select an entry from this list, the entry will be copied to the edit box. You can use the cursor keys to edit the command.
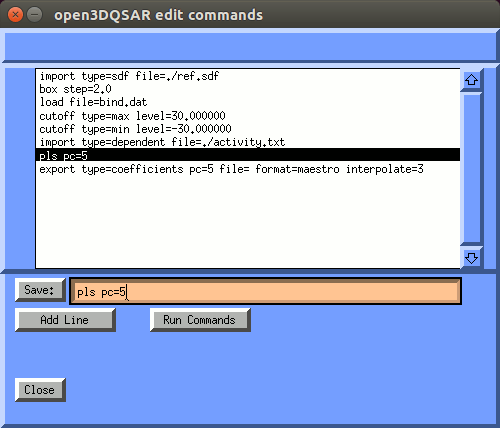
When save is clicked the edited command is saved to the list of open3dqsar commands to be executed when the "Run Commands" button is clicked.
Use environment variable OPEN3DQSAR_PATH to point to the directory where the open3dqsar executable is located. Proper operation of open3dqsar relies on the presence of openbabel, so this has to be installed as well.
The molden5.9 version comes with a full distribution for windows, Linux (64 bits ubuntu) and MacOSX (64 bits). The full distribution contains all the files necessary to run the gmolden executable, as well as the openbabel, pharmer and open3dqsar executables, with required auxilliary files.