

In the tarred source distribution of molden7.2 there are examples of
dockings in the directory test/dock.
Go to the directory
4dfr and load the file 4dfr.pdb into Molden.
Go here for a step by step recipe of docking with molden versions 7.1 and lower.
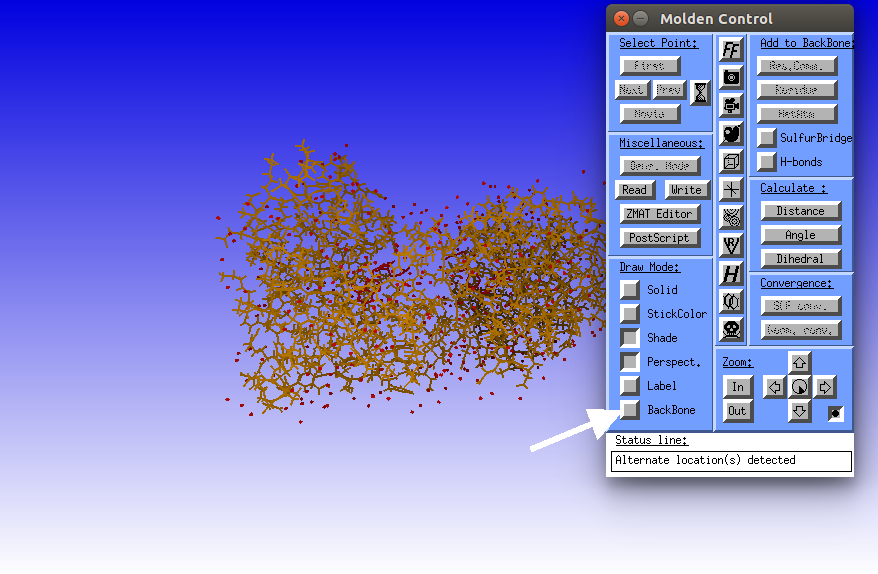
Click the BackBone button to reveal the secondary structure of the protein.
|
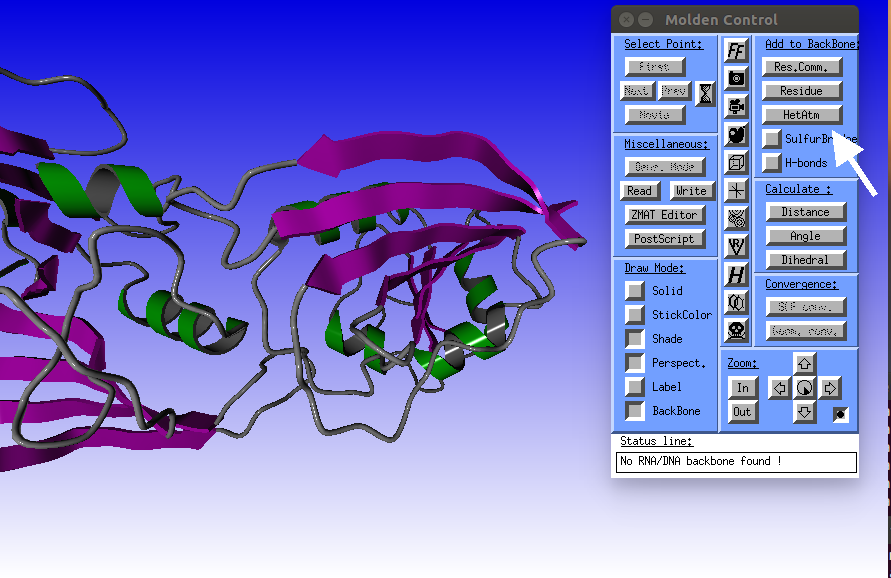
Click the HetAtm button to display the methotrexate (MTX) ligand.
|
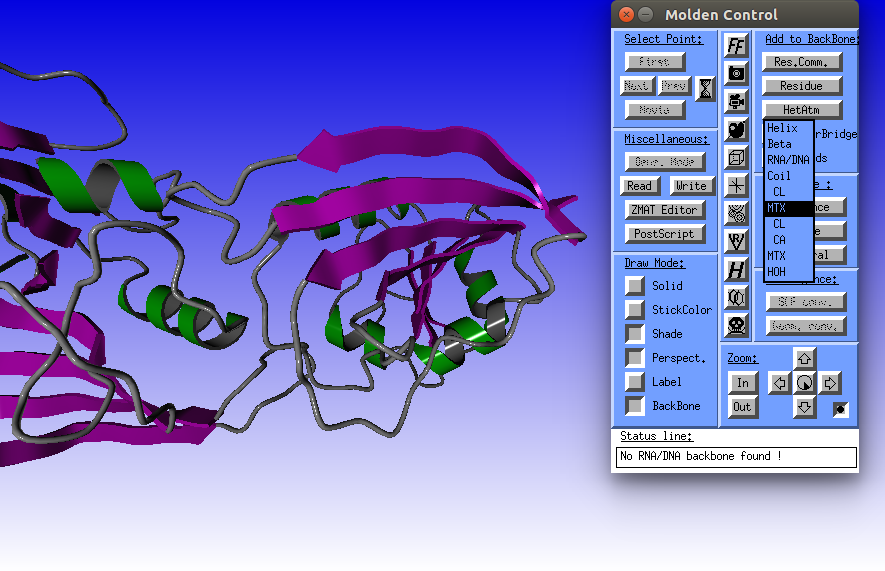
|
|
|
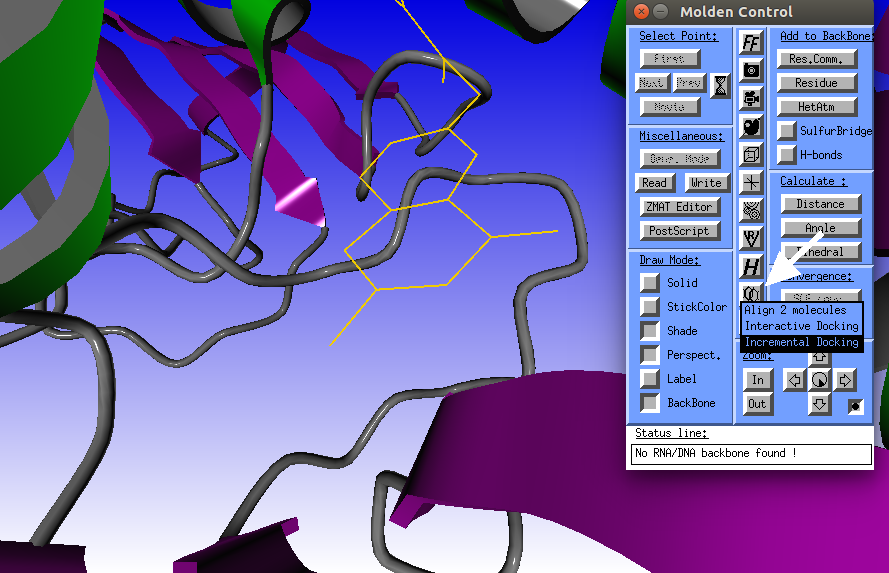
Click the "Align" icon and select the "Incremental Docking" option. |
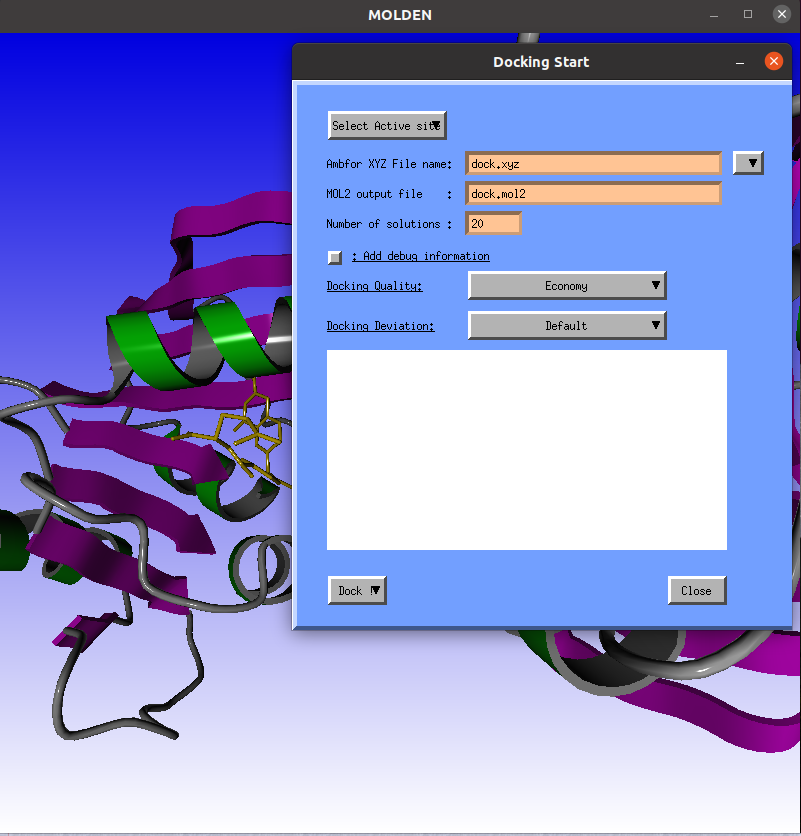
The Docking Start window will pop-up. |
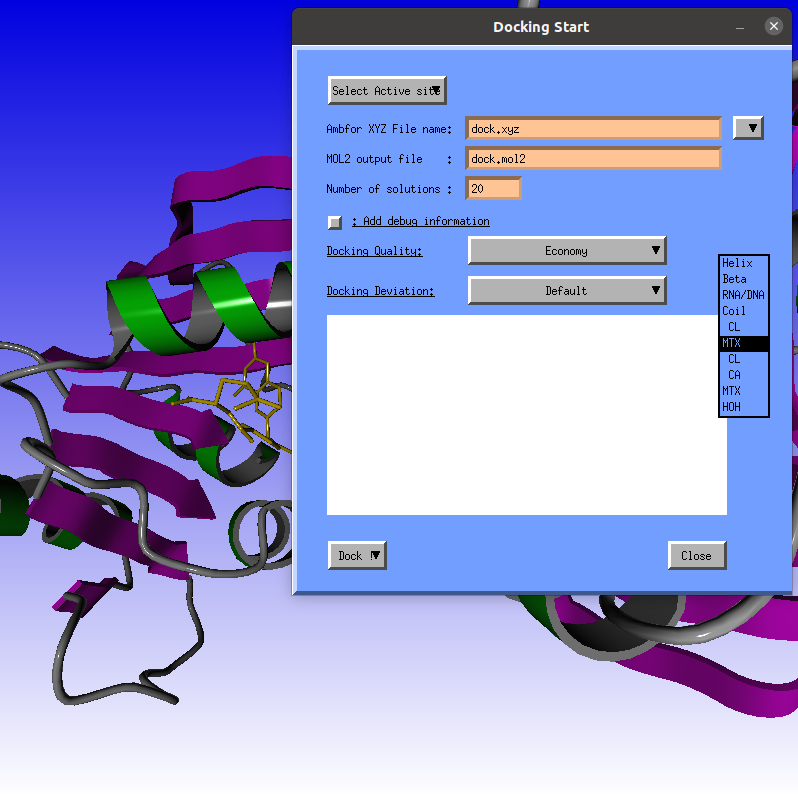
Use the first MTX record.
|
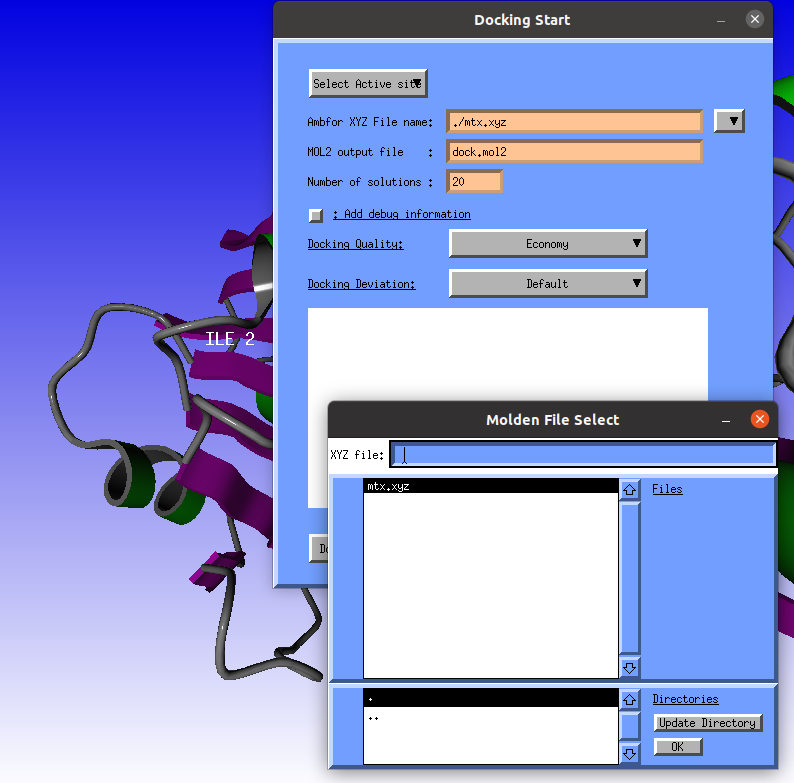
Next we need an AMBFOR .xyz file (with hydrogens) of the ligand to dock. Click the "mtx.xyz" file (contains Methotrexate) and click the OK button. |
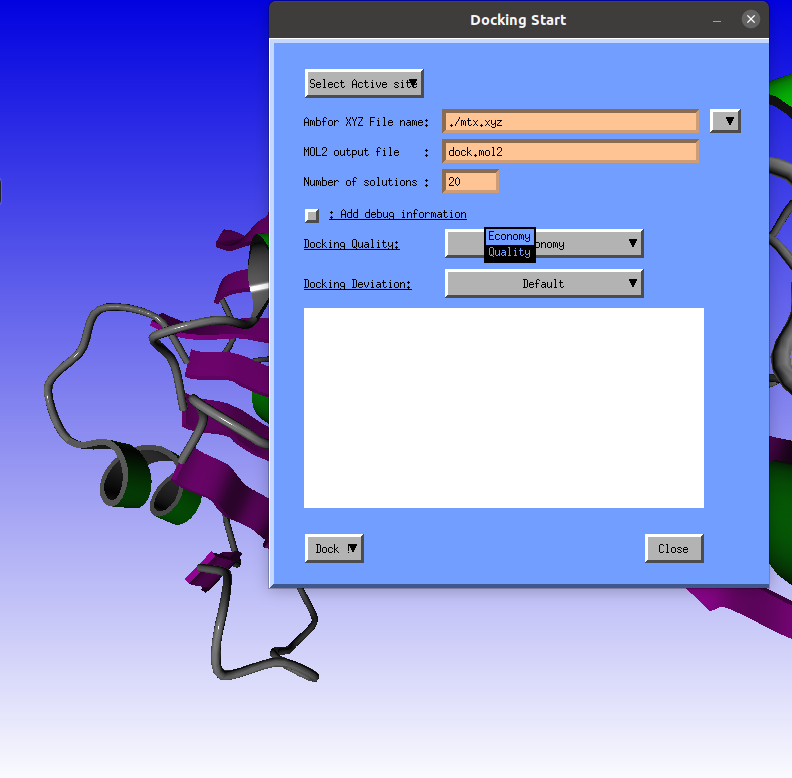
Next we need to select the "Quality" docking/ |

|
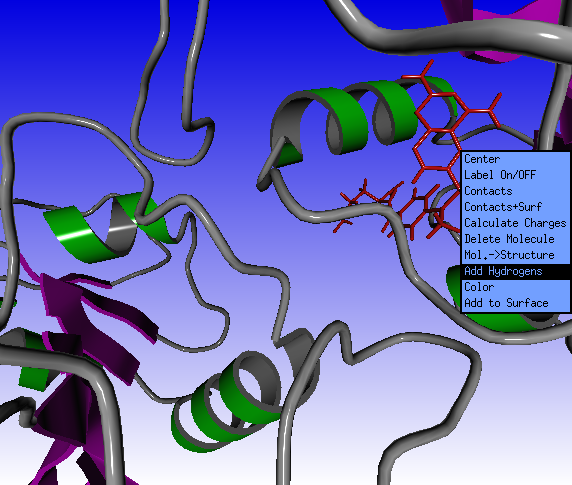
Click the Solid button in the Molden Control window and select the sticks option.
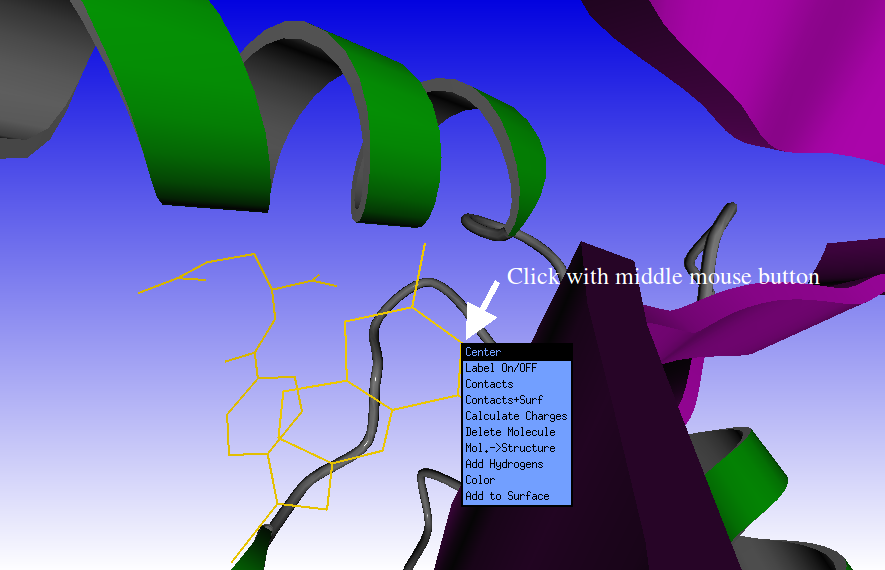
Click with the middle mouse button on the methotrexate ligand on screen
Click with the middle mouse button on the methotrexate ligand on screen
|
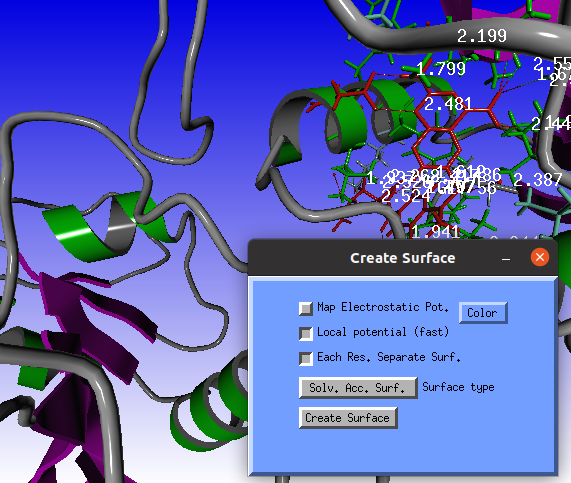
Two windows will pop-up:
|
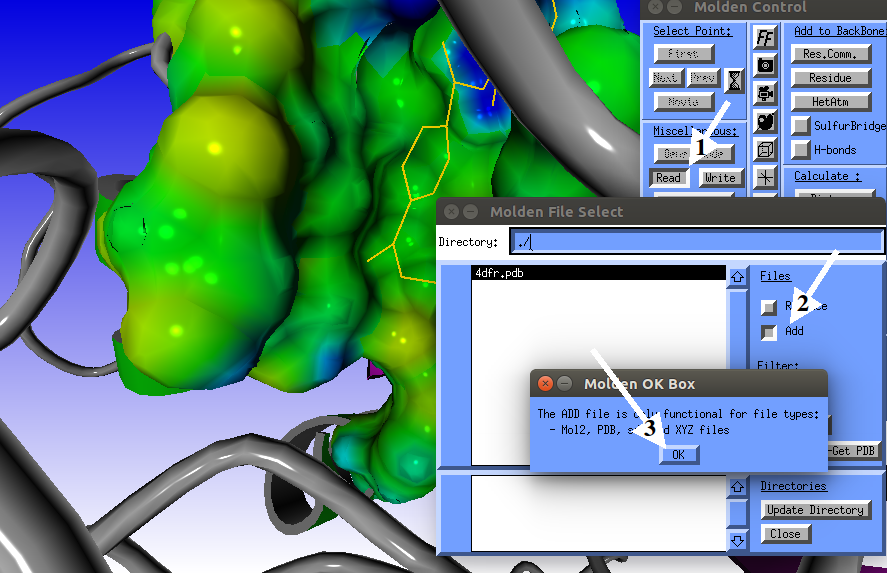
Now we are ready to read in the docking results .mol2 file.
|
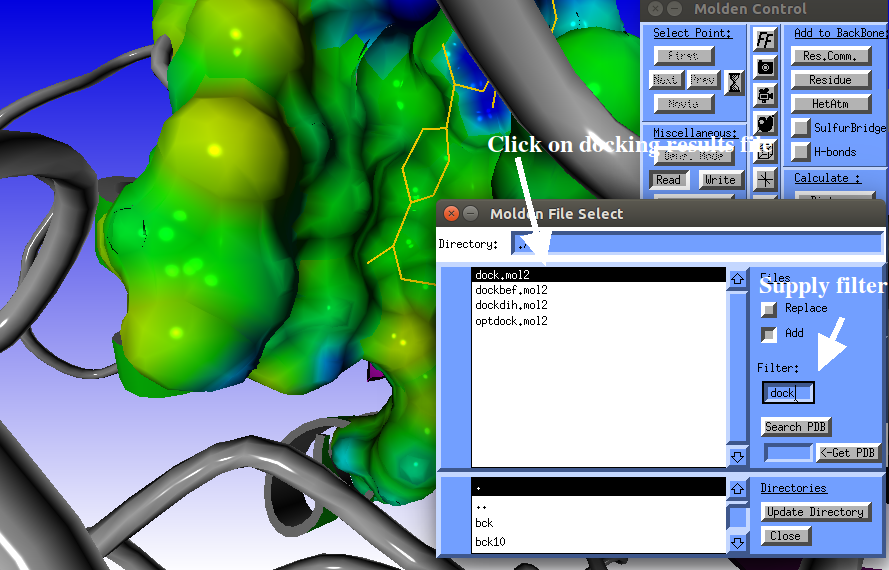
Optionally you may supply a filter, to restrict the number of files being displayed.
|
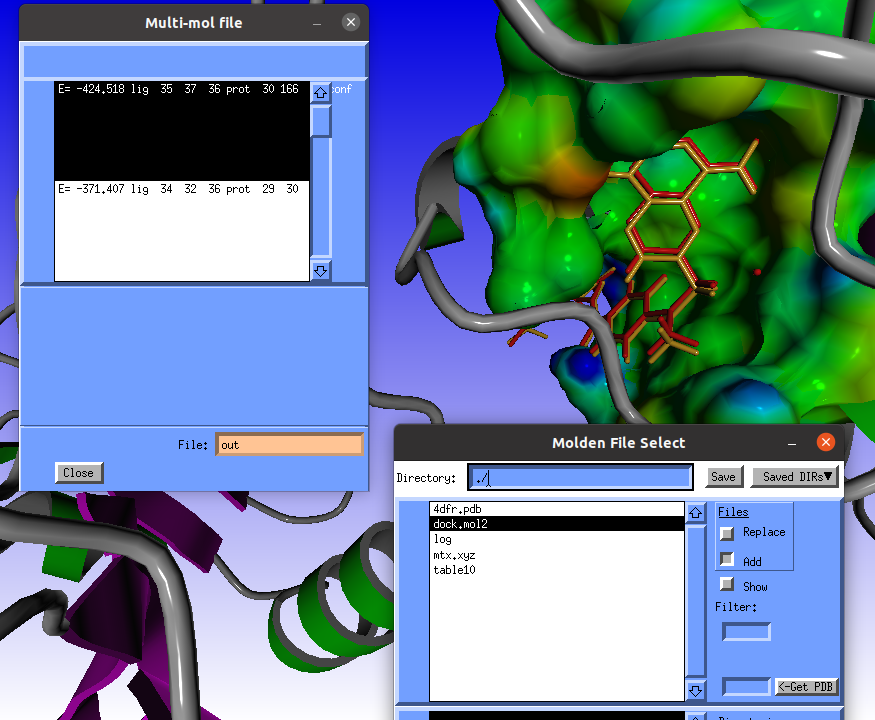
The Multi-mol file window will pop-up containg three poses:
|